Bridge Capture™
Early indexing + Linear amplification = Simplicity + Precision
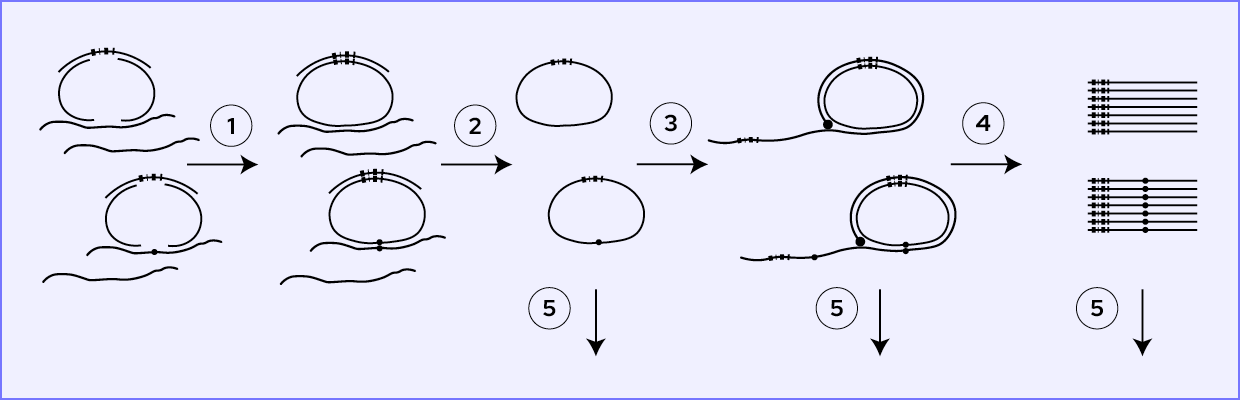
How does it perform?
Yes.
The performance of Bridge Capture™ was compared against ArcherDX LiquidPlex and AmpliSeq for Illumina Cancer Hotspot Panel v2 with comparable results. All the compared technologies were carried out by a global 3rd party CRO highly experienced with NGS.
Quote from an expert technician in NGS preparations at the CRO: “This is by far the easiest workflow I’ve ever used.”
The Bridge Capture™ workflow is designed to be fast and efficient, taking less than 5 hours to complete with only 5 minutes of hands-on time required, compared to 7 hours on amplicon-based workflows or at least 8 hours on hybrid capture-based workflows. This streamlined workflow allows for rapid sample processing, saving valuable time and resources (<See Figure 1>).
Once the workflow is complete, the subsequent NGS analysis duration depends on the sequencing technology used.

Figure 1 Duration breakdown of Bridge Capture™ Tx and MRD laboratory workflows.
The main factors that impact the sensitivity of the Bridge Capture™ are sample quality and total amount of DNA present. It is worth noting that the sensitivity varies slightly between different probes, which is most likely due to the unique binding characteristics determined by the probe and target sequences. In our publication (Adamusová et al. 2024), we observed a sensitivity range of 0.03% to 0.2% MAF, which depends on the specific probe. These MAFs were determined through serial dilutions of fragmented tumor biopsy DNA to fragmented gDNA, which were subsequently sequenced on the NovaSeq 6000. We can optimize the performance characteristics of any probes based on customer preferences.
Bridge Capture Permits Cost-Efficient, Rapid and Sensitive Molecular Precision Diagnostics
Adamusová, S., Korkiakoski, A., Hirvonen, T., Musku, A., Rantasalo, T., Laine, N., Laine, J., Blomster, J., Pursiheimo J and Tamminen, M (2024).
Patents

Accurate and massively parallel quantification of nucleic acid
- ZL201880055271.3
- EP3673081
- HK40017021
- JP7074978

Methods for accurate parallel quantification of nucleic acids in dilute or non-purified samples
- EP4060049
- US11898202

Highly sensitive methods for accurate parallel quantification of nucleic acids;
- EP4060050
- US11486003

Methods for accurate parallel detection and quantification of nucleic acids
- US11970736
