Oncology / Applications
Setting the new standard for cancer diagnostics
With Geno1® Bridge Capture™, targeted sequencing is effortless. Power up your oncological assays with industry-leading sensitivity, affordability, and scalability.
-
Affordable testing
Bridge Capture™ is engineered for cost-efficiency, utilizing off-the-shelf reagents and optimizing the use of sequencing capacity to minimize test costs.
-
Sensitive detection
Bridge Capture™ delivers proven sensitivity, meeting the rigorous demands of even the most challenging oncological applications.
-
Portable workflows
Bridge Capture™ is designed for simplicity, portability, and kittability, ensuring seamless adoption in any laboratory setting, effortless usability in distributed environments, and compatibility with any sequencing platform.
-
Pan-cancer detection
Bridge Capture™ supports panels of any size, enabling true pan-cancer detection across any number and type of mutations.
Applications
-
01
Treatment selection
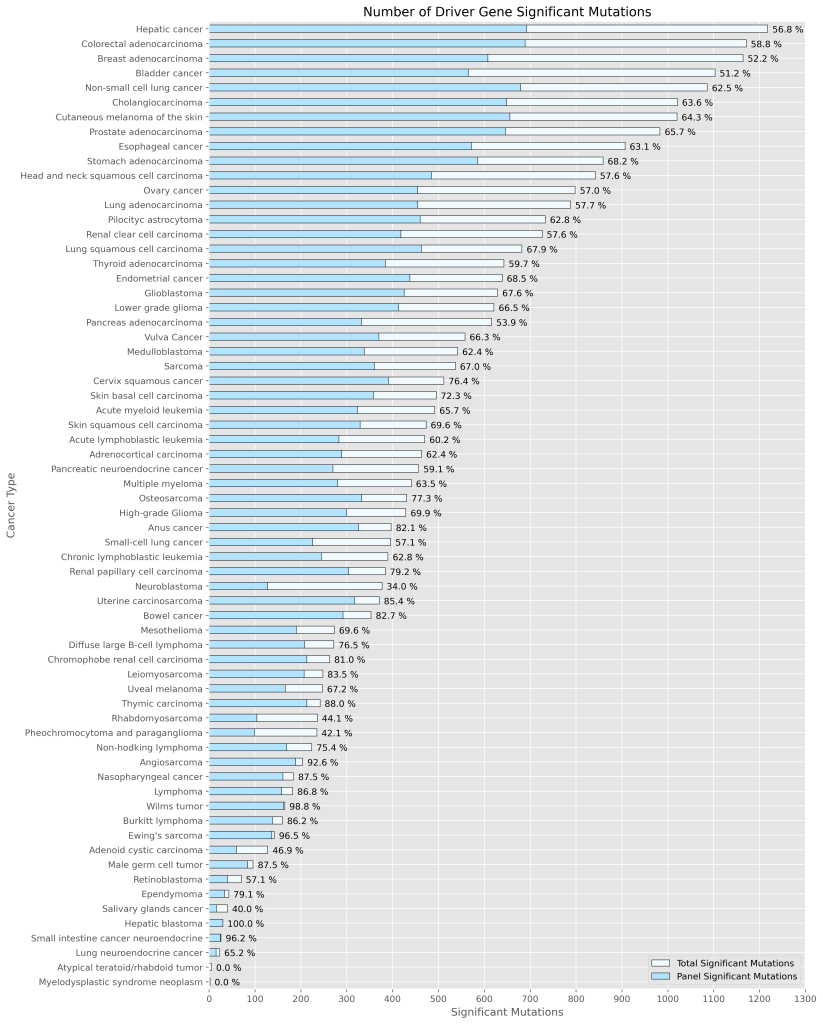
Using Genomill’s pan-cancer panel, detect up to 23’000 mutations relevant for finding the right treatment for cancer.
-
02
Minimal residual disease detection
Tumor-naive tool for monitoring cancer recurrence. Powered by Genomill’s pan-cancer panel consisting of 23’000 relevant mutations.
-
03
Early detection
Highly sensitive cancer-agnostic tool for detecting the first mutational signatures of an emerging tumor. Powered by Genomill’s pan-cancer panel consisting of 23’000 relevant mutations.
BRIDGE CAPTURE™
FAQ
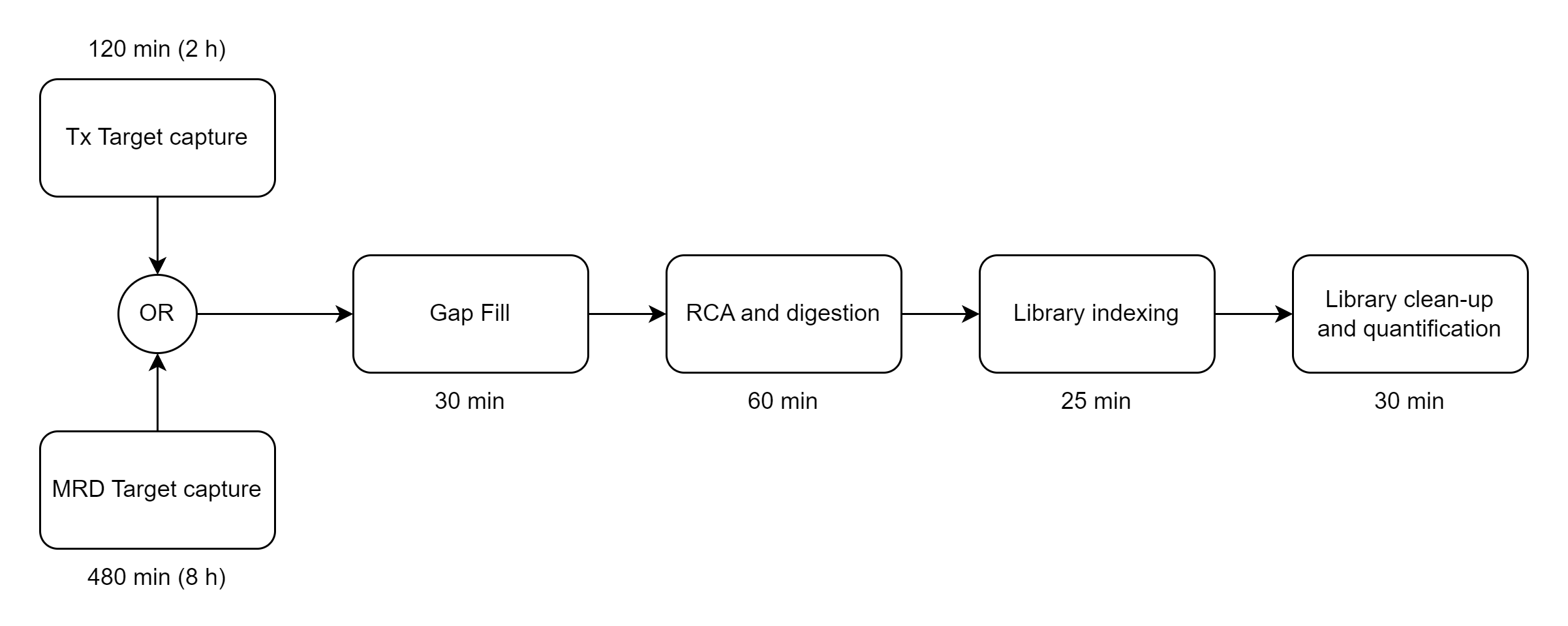
BRIDGE CAPTURE™
Panel FAQ